In Canada approximately 14,000 patients suffer from IPF [Tarride et al. 2018] mostly in older age groups (>60 years) [Hopkins et al. 2016]. The precise etiology of IPF is by definition unknown, and its diagnosis requires the exclusion of other established causes of interstitial lung disease such as autoimmune disorders and environmental exposures [Lederer & Martinez 2018]. Patients with IPF have a poor prognosis with a median survival of 3-5 years after diagnosis, accounting for approximately 1.4% of population mortality [Pleasants & Tighe 2018; Cerri et al. 2017]. In addition, the course of IPF is unpredictable, with sudden and unexplained accelerated phases of the disease (termed "exacerbations") often leading to rapid respiratory decompensation and death.
Currently, there are two anti-fibrotic drugs approved for the treatment of IPF: nintedanib and pirfenidone [Richeldi et al. 2014; King et al. 2014]. The mechanisms of action of these two drugs in IPF are poorly understood, and both have been shown to be modestly equivalent in slowing but not preventing the progressive decline of lung function [Flaherty et al. 2019]. Due to its relentless progression and the limited therapeutic options, new approaches to IPF treatment are desperately needed.
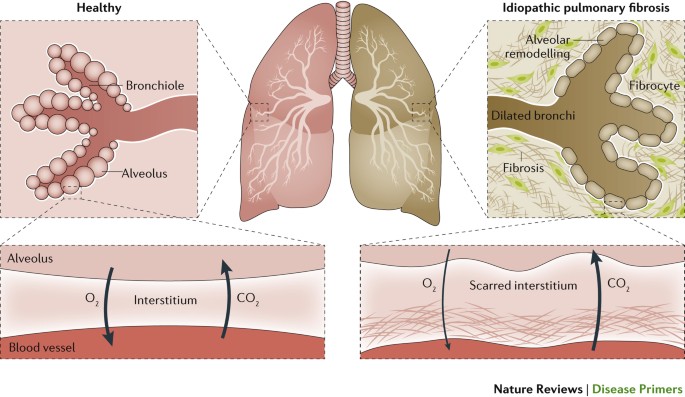
There is evidence in mouse models that indicate an important role for lung macrophages in the induction and maintenance of the fibrotic phenotype. This phenotype is likely driven by a combination of aging lung resident macrophages driving inappropriate inflammation, a process called inflamm-aging, and an increase in monocyte-derived macrophages which do not properly respond to insult [McCubbrey et al. 2018; Ucero et al. 2019; Reyfman et al. 2019].
Our research program uses lung macrophages to study the profibrotic environment of the lung in IPF and the changes macrophages undergo in aging and IPF. Additionally, we use peripheral monocytes to define biomarkers of IPF for diagnosis and prognosticators of response to anti-fibrotic drugs for treatment and predictors of acute exacerbation.
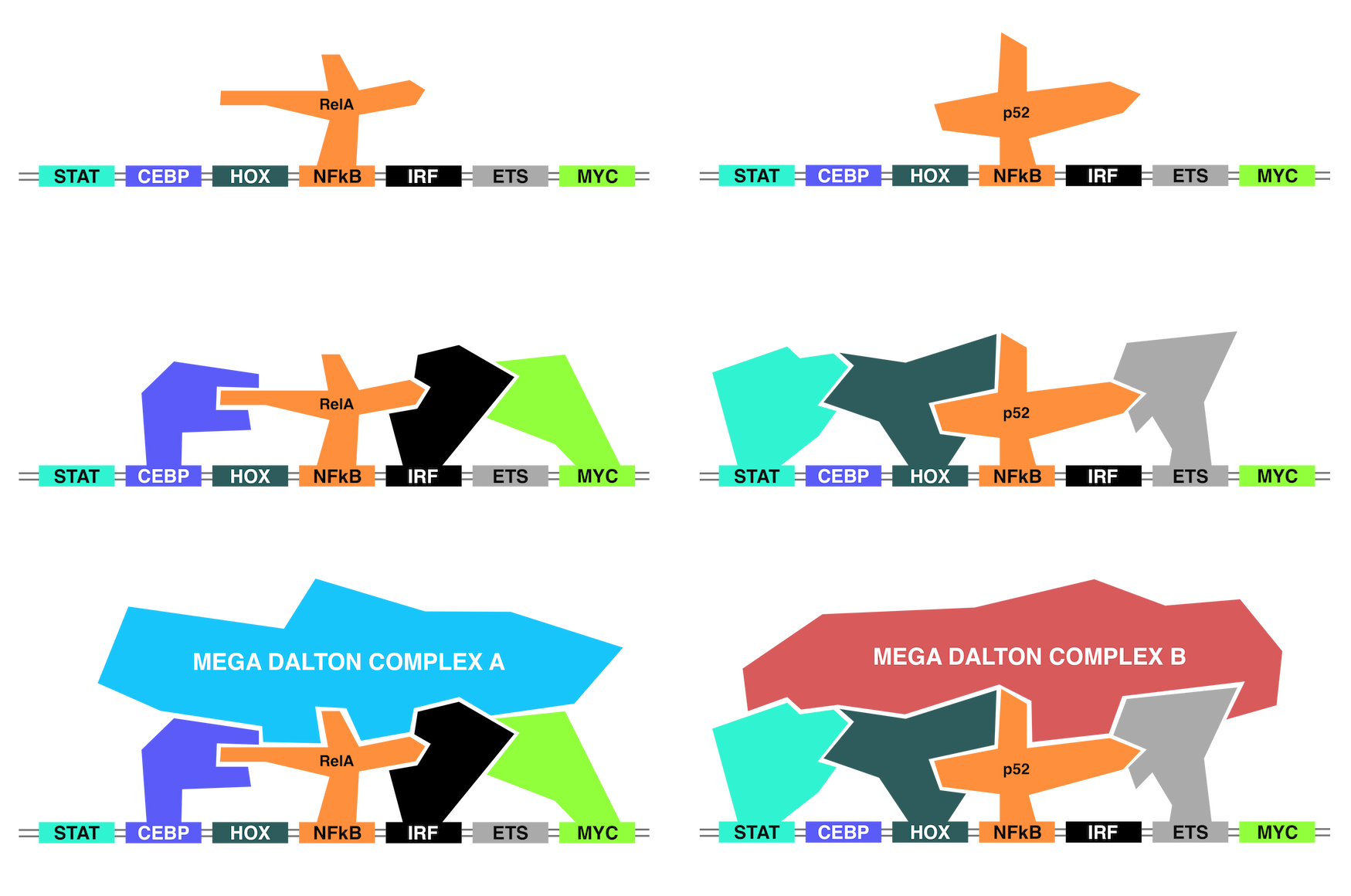
The DNA is a 4-letter language that provides all of the information to locate and express genes. The targeting and expression of each gene is performed through the recruitment of proteins called transcription factors (TFs). Each TF recognizes and is recruited to the genome by a specific code, called a DNA binding motif (DBM) [Khan et al. 2018; Grant et al. 2011; Bailey et al. 2009]. Like a lock and key, TFs target regulatory elements through a DNA binding domain to a compatible DNA element [Zhang et al. 2017; Fonseca & Tao et al. 2019; Halazonetis et al. 1988]. The consensus sequence for each DNA element is called a DNA binding motif.
However, the presence of one DBM is not sufficient to recruit a TF as DNA motifs are common throughout the genome (millions of sites), while the binding of TFs is relatively rare (thousand to tens of thousands of sites). Instead, TFs must work in coordinated groups to cooperatively bind to DNA regions and activate transcription. This means that the underlying DNA must contain the DNA binding motifs of these coordinated sets of TFs for transcriptional activation or repression [Kaikkonen et al. 2013; Heinz et al. 2010]. The TF sets then interpret the underlying DNA and coordinate the binding of chromatin modifiers, coactivators and coregulatory factors that must fit together in a large and intricate puzzle that provides the appropriate output on a gene [Liu et al. 2014].
The interpretation of DNA is further complicated by TF families which may contain dozens of family members capable of binding to similar if not identical DNA binding motifs. In these circumstances, the surrounding TFs are responsible for determining the TF family member specificity [Heinz et al. 2013; Barozzi et al. 2014]. The underlying DNA code provides the rules of DNA binding; the DNA motifs recruit specific TFs through their DNA binding domains. Then, the TFs capable of binding select different TF family members based on compatibility [Wei et al. 2010; Curina et al. 2017].
In this work, we use machine learning models to tease apart the genomic language by understanding how the cell reads the DNA to perform transcriptional activation.